From docs/XSBench_Theory:
The XSBench proxy app models the most computationally intensive part of a typical MC transport algorithm – the calculation of macroscopic neutron cross sections – a kernel which accounts for around 85% of the total runtime of OpenMC. The essential computational conditions and tasks of fully featured MC neutron transport codes are retained in the mini-app, without the additional complexity of the full application.
Problem Size and Run Configuration
Version 18:
The work which XSBench proxies is the cross-section lookups, in version 18 this is best controlled by the -p <particles> parameter. Increasing this value will increase the amount of work for the proxy (number of particles X number of lookups per particle).
Problems with 10^9 particles are not unusual for a current run. Scaling the number of particles beyond 10^12 will not likely be meaningful.
Analysis
Build and Run Information – Version 16
Compiler = icc (ICC) 18.0.1 20171018
Build Flags = -g -O3 -march=native -qopenmp
Run Parameters = -s large -g 11303 -G unionized -l 100000000 -t [#ofThreads]
Version 16 of XSBench scaled its workload with number of lookups (-l) instead of number of particles X number of lookups.
Scaling
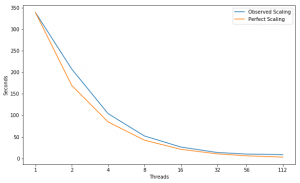
Performance Improvement
| Threads | |||||||
|---|---|---|---|---|---|---|---|
| Speed Up | 1.63X | 2.00X | 2.00X | 1.97X | 1.92X | 1.37X | 1.13X |
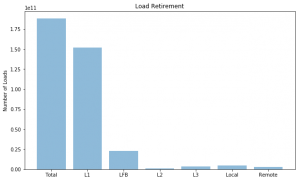
Hit Locations
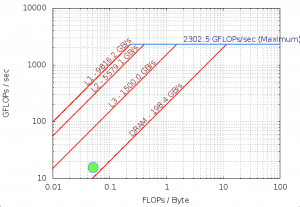
FLOPS
| Double Precision | ||||||
|---|---|---|---|---|---|---|
| PMU | 4.720e+10 | 0.000e+00 | 2.790e+10 | 0.000e+00 | 1.588e+11 | 1.574e+01 |
| SDE | 4.584e+10 | 0.000e+00 | 2.222e+10 | 0.000e+00 | 1.347e+11 | 1.328e+01 |
Intel Software Development Emulator
| Intel SDE | |
|---|---|
| Arithmetric Intensity | 0.0515 |
| FLOPS per Inst | 0.286 |
| FLOPS per FP Inst | 1.98 |
| Bytes per Load Inst | 10.3 |
| Bytes per Store Inst | 11.8 |
Roofline – Intel(R) Xeon(R) Platinum 8180M CPU
112 Threads – 56 – Cores 3200.0 Mhz
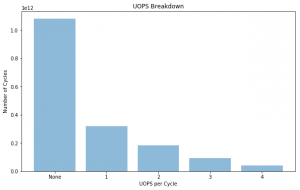
UOPS Executed
Experiment Aggregate Metrics
| 1 (100.0%) | 0.54 | 0.43 | 0.34 | 7.10% | 46.89% | 68.53% | 4.04% | 13.22% | 14.63% |
| 56 (100.0%) | 0.35 | 0.27 | 0.22 | 6.86% | 46.25% | 66.82% | 2.33% | 15.89% | 55.09% |
| 112 (100.0%) | 0.40 | 0.16 | 0.13 | 6.91% | 44.55% | 67.22% | 2.69% | 17.60% | 58.67% |
main omp loop
153 // OpenMP compiler directives - declaring variables
// as shared or private
154 #pragma omp parallel default(none) \
155 private(i, thread, p_energy, mat, seed) \
156 shared( in, energy_grid, nuclide_grids, \
157 mats, concs, num_nucs, mype, vhash)
158 {
159 // Initialize parallel PAPI counters
160 #ifdef PAPI
{...}
167 #endif
168
169 double macro_xs_vector[5];
170 double * xs = (double *) calloc(5, sizeof(double));
171
172 // Initialize RNG seeds for threads
173 thread = omp_get_thread_num();
174 seed = (thread+1)*19+17;
175
176 // XS Lookup Loop
177 #pragma omp for schedule(dynamic)
178 for( i = 0; i < in.lookups; i++ )
179 {
180 // Status text
181 if( INFO && mype == 0 &&
thread == 0 && i % 1000 == 0 )
182 printf("\rCalculating XS's...
(%.0lf%% completed)",
183 (i / ( (double)in.lookups / (double) in.nthreads ))
184 / (double) in.nthreads * 100.0);
185
186 // Randomly pick an energy and material
// for the particle
187 #ifdef VERIFICATION
{...}
196 #endif
197
198 // debugging
199 //printf("E = %lf mat = %d\n", p_energy, mat);
200
201 // This returns the macro_xs_vector, but we're not going
202 // to do anything with it in this program, so return value
203 // is written over.
204 calculate_macro_xs( p_energy, mat,
in.n_isotopes,
205 in.n_gridpoints,
num_nucs, concs,
206 energy_grid,
nuclide_grids, mats,
207 macro_xs_vector,
in.grid_type,
in.hash_bins );
208
209 // Copy results from above function call onto heap
210 // so that compiler cannot optimize function out
211 // (only occurs if -flto flag is used)
212 memcpy(xs, macro_xs_vector, 5*sizeof(double));
213
214 // Verification hash calculation
215 // This method provides a consistent hash accross
216 // architectures and compilers.
217 #ifdef VERIFICATION
{...}
229 #endif
230 }
231
232 // Prints out thread local PAPI counters
233 #ifdef PAPI
{...}
246 #endif
248 }
calculate_macro_xs
| 1 (97.4%) | 0.50 | 0.41 | 0.33 | 7.49% | 46.83% | 68.69% | 4.04% | 13.31% | 14.86% |
| 56 (83.4%) | 0.36 | 0.28 | 0.23 | 7.31% | 46.53% | 67.07% | 2.69% | 18.55% | 64.65% |
| 112 (72.6%) | 0.44 | 0.19 | 0.15 | 7.36% | 44.90% | 67.44% | 3.30% | 21.73% | 72.17% |
101 void calculate_macro_xs( double p_energy, int mat, long n_isotopes,
102 long n_gridpoints, int * restrict num_nucs,
103 double ** restrict concs,
104 GridPoint * restrict energy_grid,
105 NuclideGridPoint ** restrict nuclide_grids,
106 int ** restrict mats,
107 double * restrict macro_xs_vector,
int grid_type, int hash_bins ){
108 double xs_vector[5];
109 int p_nuc; // the nuclide we are looking up
110 long idx = -1;
111 double conc; // the concentration of the nuclide
// in the material
112
113 // cleans out macro_xs_vector
114 for( int k = 0; k < 5; k++ )
115 macro_xs_vector[k] = 0;
116
117 // If we are using the unionized energy grid (UEG), we only
118 // need to perform 1 binary search per macroscopic lookup.
119 // If we are using the nuclide grid search, it will have to be
120 // done inside of the "calculate_micro_xs" function for each
// different
121 // nuclide in the material.
122 if( grid_type == UNIONIZED )
123 idx = grid_search( n_isotopes * n_gridpoints, p_energy,
124 energy_grid);
125 else if( grid_type == HASH )
126 {
{...}
129 }
130
131 // Once we find the pointer array on the UEG, we can pull the data
132 // from the respective nuclide grids, as well as the nuclide
133 // concentration data for the material
134 // Each nuclide from the material needs to have
// its micro-XS array
135 // looked up & interpolatied (via calculate_micro_xs). Then, the
136 // micro XS is multiplied by the concentration of that nuclide
137 // in the material, and added to the total macro XS array.
138 for( int j = 0; j < num_nucs[mat]; j++ )
139 {
140 p_nuc = mats[mat][j];
141 conc = concs[mat][j];
142 calculate_micro_xs( p_energy, p_nuc, n_isotopes,
143 n_gridpoints, energy_grid,
144 nuclide_grids, idx, xs_vector,
grid_type, hash_bins );
145 for( int k = 0; k < 5; k++ )
146 macro_xs_vector[k] += xs_vector[k] * conc;
147 }
{...}
155 }
[I] grid_search
| 1 (16.3%) | 0.29 | 0.06 | 0.04 | 38.37% | 38.04% | 37.93% | 4.48% | 12.55% | 14.54% |
| 56 (12.8%) | 0.23 | 0.05 | 0.03 | 38.00% | 38.60% | 35.61% | 3.22% | 19.44% | 68.60% |
| 112 (7.9%) | 0.41 | 0.04 | 0.02 | 43.16% | 39.89% | 35.28% | 4.74% | 28.81% | 96.22% |
158 // (fixed) binary search for energy on unionized energy grid
159 // returns lower index
160 long grid_search( long n, double quarry, GridPoint * A)
161 {
162 long lowerLimit = 0;
163 long upperLimit = n-1;
164 long examinationPoint;
165 long length = upperLimit - lowerLimit;
166
167 while( length > 1 )
168 {
169 examinationPoint = lowerLimit + ( length / 2 );
170
171 if( A[examinationPoint].energy > quarry )
172 upperLimit = examinationPoint;
173 else
174 lowerLimit = examinationPoint;
175
176 length = upperLimit - lowerLimit;
177 }
178
179 return lowerLimit;
180 }
calculate_micro_xs
| 1 (72.2%) | 0.49 | 0.42 | 0.35 | 6.11% | 49.46% | 72.74% | 3.43% | 11.42% | 14.36% |
| 56 (65.5%) | 0.34 | 0.28 | 0.24 | 5.83% | 48.97% | 72.66% | 2.37% | 16.55% | 61.84% |
| 112 (61.0%) | 0.42 | 0.18 | 0.15 | 6.39% | 45.93% | 72.88% | 2.95% | 19.66% | 68.11% |
3 // Calculates the microscopic cross section for a given nuclide & energy
4 void calculate_micro_xs( double p_energy, int nuc, long n_isotopes,
5 long n_gridpoints,
6 GridPoint * restrict energy_grid,
7 NuclideGridPoint ** restrict nuclide_grids,
8 long idx, double * restrict xs_vector, int grid_type,
int hash_bins ){
9 // Variables
10 double f;
11 NuclideGridPoint * low, * high;
12
13 // If using only the nuclide grid, we must perform a binary search
14 // to find the energy location in this particular nuclide's grid.
15 if( grid_type == NUCLIDE )
16 {
{...}
26 }
27 else if( grid_type == UNIONIZED) // Unionized Energy Grid - we already know
// the index, no binary search needed.
28 {
29 // pull ptr from energy grid and check to ensure that
30 // we're not reading off the end of the nuclide's grid
31 if( energy_grid[idx].xs_ptrs[nuc] == n_gridpoints - 1 )
32 low = &nuclide_grids[nuc][energy_grid[idx].xs_ptrs[nuc] - 1];
33 else
34 low = &nuclide_grids[nuc][energy_grid[idx].xs_ptrs[nuc]];
35 }
36 else // Hash grid
37 {
{...}
65 }
66
67 high = low + 1;
68
69 // calculate the re-useable interpolation factor
70 f = (high->energy - p_energy) / (high->energy - low->energy);
71
72 // Total XS
73 xs_vector[0] = high->total_xs - f * (high->total_xs - low->total_xs);
74
75 // Elastic XS
76 xs_vector[1] = high->elastic_xs - f * (high->elastic_xs
- low->elastic_xs);
77
78 // Absorbtion XS
79 xs_vector[2] = high->absorbtion_xs - f * (high->absorbtion_xs
- low->absorbtion_xs);
80
81 // Fission XS
82 xs_vector[3] = high->fission_xs - f * (high->fission_xs
- low->fission_xs);
83
84 // Nu Fission XS
85 xs_vector[4] = high->nu_fission_xs - f * (high->nu_fission_xs
- low->nu_fission_xs);
86
{...}
98 }